Original article published in NIH National Library of Medicine
(Structured Abstract)
INTRODUCTION
For much of modern human history, we were only one of several different groups of hominins that existed. Studies of ancient and modern DNA have shown that admixture occurred multiple times among different hominin lineages, including between the ancestors of modern humans and Neanderthals. A number of methods
have been developed to identify Neanderthal-introgressed sequences in the DNA of modern humans, which have provided insight into how admixture with Neanderthals shaped the biology and evolution of modern human genomes. Although gene flow from an early modern human population to Neanderthals has been described, the consequences of admixture on the Neanderthal genome have received comparatively less attention.
RATIONALE
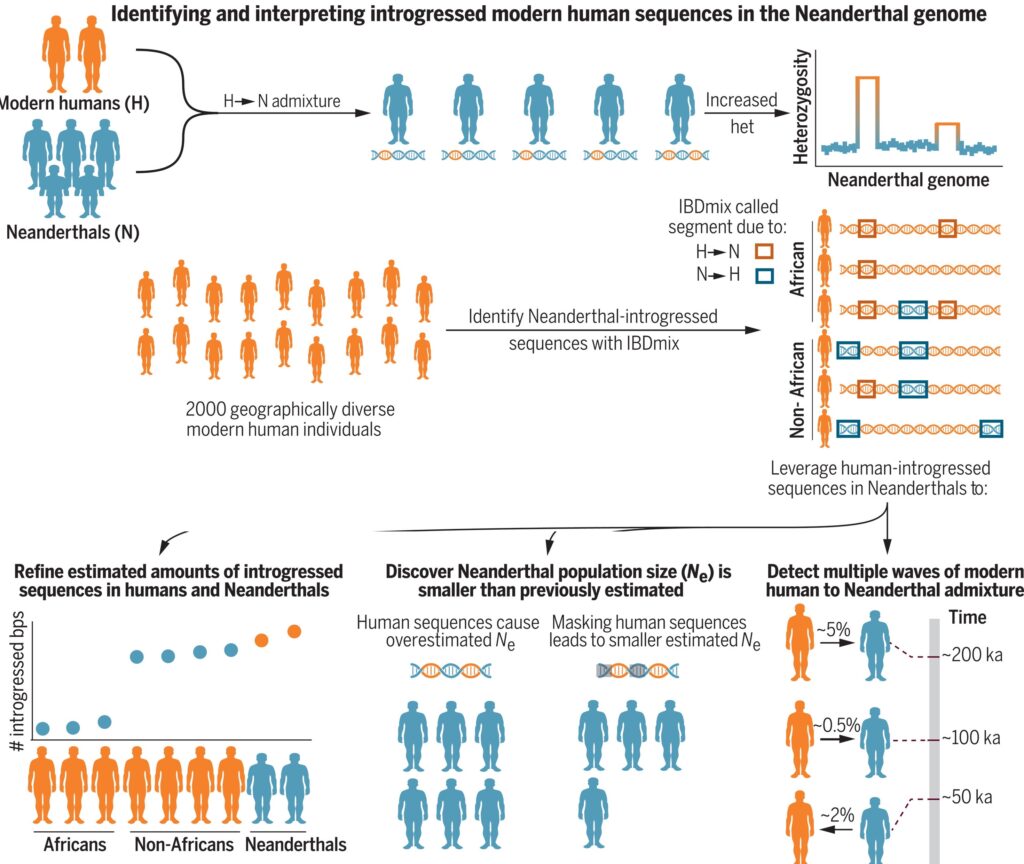
A better understanding of how admixture with modern humans influenced patterns of Neanderthal genomic variation may provide insights into hominin evolutionary history. For example, DNA sequences inherited from modern human ancestors in Neanderthals can be used to test hypotheses on the frequency, magnitude, and timing of admixture and the population genetics characteristics of Neanderthals. Introgressed modern human sequences in Neanderthals can also be used to refine estimates of Neanderthal ancestry in contemporary individuals. We developed a simple framework to investigate introgressed human sequences in Neanderthals that is predicated on the expectation that sequences inherited from modern human ancestors would be, on average, more genetically diverse and would result in local increases in heterozygosity across the Neanderthal genome.
RESULTS
We first used a method referred to as IBDmix to identify introgressed Neanderthal sequences in 2000 modern humans sequenced by the 1000 Genomes Project. We
found that sequences identified by IBDmix as Neanderthal in African individuals from the 1000 Genomes Project are significantly enriched in regions of high heterozygosity in the Neanderthal genome, whereas no such enrichment is observed with sequences detected as introgressed in non-African individuals. We show that these patterns are caused by gene flow from modern humans to Neanderthals and estimate that the Vindija and Altai Neanderthal genomes have 53.9 Mb (2.5%) and 80.0 Mb (3.7%) of human-introgressed sequences, respectively. We leverage human-introgressed sequences in Neanderthals to revise estimates of the amount of Neanderthal-introgressed sequences in modern humans. Additionally, we show that human-introgressed sequences cause Neanderthal population size to be overestimated and that accounting for their effects decrease estimates of Neanderthal population size by ~20%. Finally, we found evidence for two distinct epochs of human gene flow into Neanderthals.
CONCLUSION
Our results provide insights into the history of admixture between modern humans and Neanderthals, show that gene flow had substantial impacts on patterns of modern human and Neanderthal genomic variation, and show that accounting for human-introgressed sequences in Neanderthals enables more-accurate inferences of admixture and its consequences in both Neanderthals and modern humans. More generally, the smaller estimated population size and inferred admixture dynamics are consistent with a Neanderthal population that was decreasing in size over time and was ultimately being absorbed into the modern human gene pool.